之前试了文献提供的去除批次效应的代码,发现效果非常差,还是用标准流程走一下看看吧
读入数据
1
2
3
4
5
6
7
| library(scran);
library(tidyverse);
library(Seurat);
sce <- readRDS('scRNA.rds')
sce <- as.Seurat(sce)
sceL <- SplitObject(object = sce, split.by = 'Batch')
table(sce[['Batch']])
|
分开的10x数据文件夹
1
2
3
4
5
6
7
8
9
10
11
12
13
14
| root_path = "."
f_read10x <- function(dirN, ...){
tp_samples <- list.files(file.path(root_path, dirN))
tp_dir <- file.path(root_path, dirN, tp_samples)
names(tp_dir) <- tp_samples
scRNA <- list()
for (lc_ba in names(tp_dir)){
counts <- Read10X(data.dir = tp_dir[[lc_ba]])
scRNA[[lc_ba]] <- CreateSeuratObject(counts, project = lc_ba, ...)
scRNA[[lc_ba]][["percent.mt"]] <- PercentageFeatureSet(scRNA[[lc_ba]], pattern = "^MT-")
scRNA[[lc_ba]][["percent.ERCC"]] <- PercentageFeatureSet(scRNA[[lc_ba]], pattern = "^ERCC-")
}
scRNA
}
|
标准整合流程
1
2
3
4
5
|
sceL <- lapply(X = sceL, FUN = function(x){FindVariableFeatures(x, selection.method = "vst", nfeatures = 2000)})
sceL.anchors <- FindIntegrationAnchors(object.list = sceL, dims = 1:30)
sceL.integrated <- IntegrateData(anchorset = sceL.anchors, dims = 1:30)
saveRDS(sceL.integrated, 'sceL.integrated.rds')
|
最终效果
1
2
3
4
| sce <- as.SingleCellExperiment(sceL.integrated)
library(scater);
options(repr.plot.width=7, repr.plot.height=6)
plotTSNE(sce, colour_by="Batch")
|
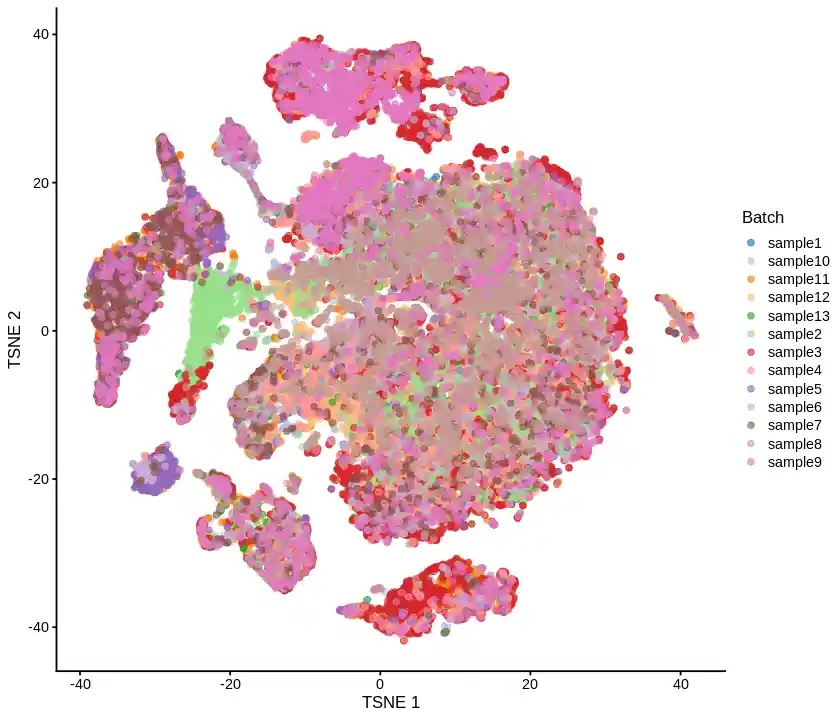
emm,效果差强人意

